Alpha satellite DNA: Difference between revisions
(Created page) |
(No difference)
|
Revision as of 07:28, 28 January 2023
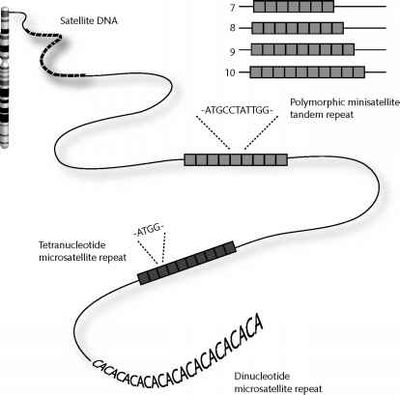
Alpha satellite DNA is a tandemly organized type of repetitive DNA that comprises 5% of the genome and is found at all human centromeres. A defined number of 171-bp monomers are organized into chromosome-specific higher-order repeats (HORs) that are reiterated thousands of times.
Satellite DNA comprises very large arrays of tandemly repeated noncoding DNA, usually megabases in size, which form the major structural constituent of hetero-chromatin, a tightly packaged condensed state of transcriptionally suppressed DNA.
Satellite DNA is the main component of functional centromeres, as well as heterochromatin in pericentromeric and telomeric regions, and in the short arms of the acrocentric chromosomes (chromosomes 13, 14, 15, 21, and 22) (Charlesworth et al. 1994; Schueler et al. 2001). Satellite DNA is recognized across eukaryotic genomes in some species, such as the kangaroo rat Dipodomys ordii comprising half of the total genomic DNA content (Hatch and Mazrimas 1974).
The technical challenges of sequencing and analysing such large repetitive DNA sequences have meant only limited coverage in genome assemblies. Our understanding of the precise structure, organization, and functional importance of satellite DNA still needs to be completed.
The unusual structure of satellite DNA may be the preferred state at centromeres and flanking regions. The specific composition of satellite DNA is variable and typically species-specific.
Concerted evolution is thought to maintain sequence homogeneity of species-specific satellite arrays but allows for the rapid change in sequence or composition of repeats required during evolution (Plohl et al. 2008).
(Source: https://www.fossilhunters.xyz/genetic-diversity/satellite-dna.html)